| Package |
|---|
| base |
| gt |
| here |
| janitor |
| knitr |
| palmerpenguins |
| rmarkdown |
| skimr |
| tidyverse |
Crash Course in R
Princeton University
2023-09-10
Packages
Objective
Get you familiar with R
Understand basic terminology and concepts
Learn how to run commands
Manipulate data
Load and save data/scripts/QMD/RMD

It’s scary

Note
You can learn R!
You will get frustrated
You will get errors that don’t help or make sense
- Google is your friend
- Try googling the specific error message first
- Then try googling your specific function and the error
- Try a bunch of different search terms.
- ChatGPT (use wisely)
- Me :)
- Google is your friend
Outline
Why RIDE
R commands, data structures, and functions
Tidyverse & the pipe operator
Reading in data
Manipulating data
Saving data
Why R?
- Free and open-source
- Flexibility
- Programming language (not point-and-click)
- Excellent graphics (via
ggplot2)
- Easy to generate reproducible reports (markdown and quarto)
- Easy to integrate with other tools and programs
- Inclusive community
- Marketability
Outline
Why R?
IDER commands, data structures, and functions
Tidyverse & the pipe operator
Multiple functions
Reading in data
Saving R scripts
RStudio IDE
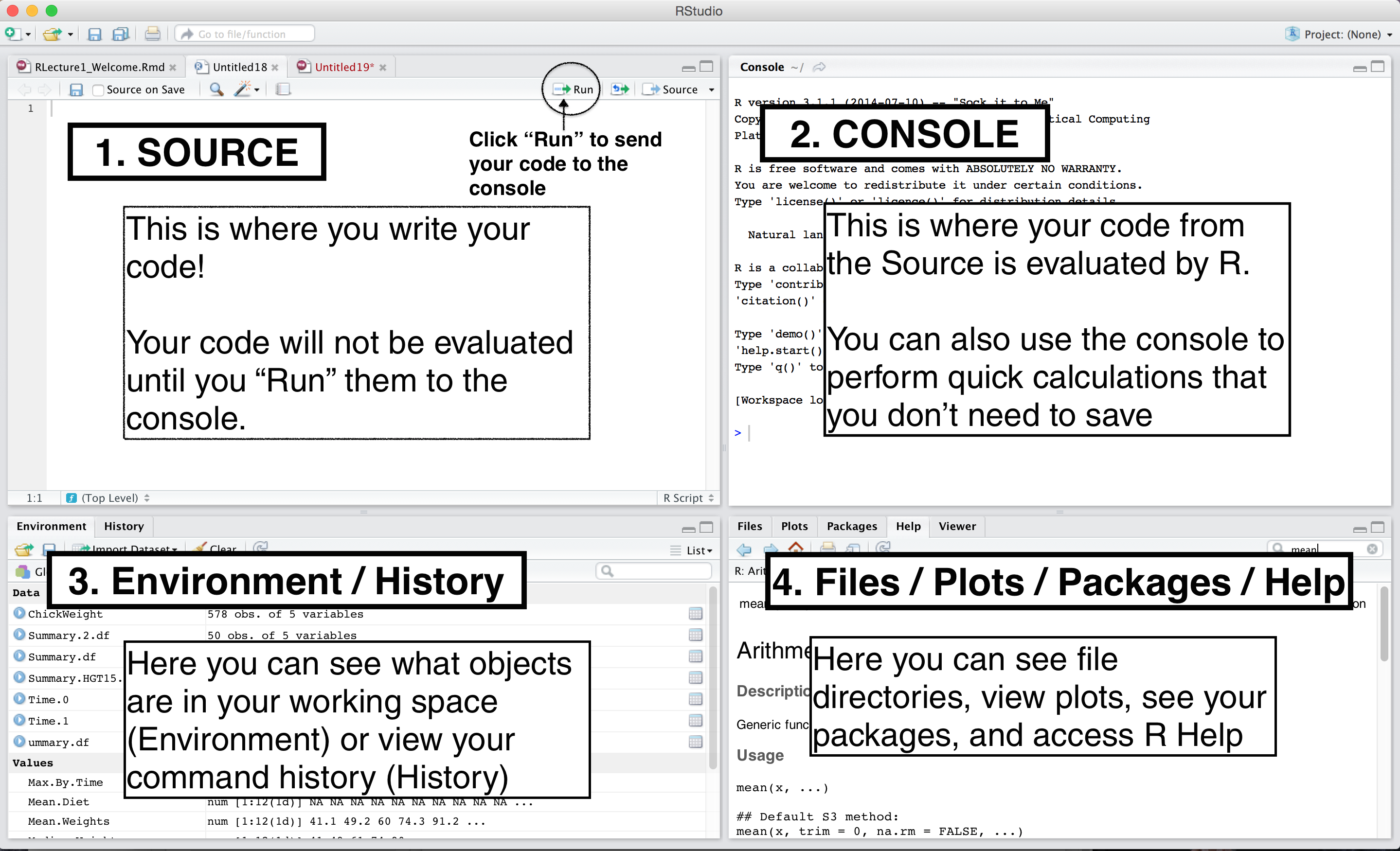
RStudio IDE
- Make it your own!

Outline
Why R?
RStudio
R commands, data structures, and functionsTidyverse & the pipe operator
Reading in data
Saving R scripts
Commands
Commands are the code that you tell R to do for you
They can be very simple or complex
Computers do what you tell them to do. Mistakes happen!
- Maybe it’s a typo, maybe it’s a misunderstanding of what the code does
You can type a command directly into the console
You can type in a document (Script or Markdown/Quarto) and tell it to then run in the console
Commands
>indicates the console is ready for more code+indicates that you haven’t finished a code blockCapitalization and symbols matter
Hit the up arrow – you can scroll through the last commands that were run
Hit the tab key – you’ll get a list of variable names and options to select from
Use the
?followed by a command to learn more about it
Assignment
- Few different way to assign values to objects
The arrow
<-is called an ASSIGNMENT OPERATOR, and tells R to save an object called x that has the value of 4- Even
->(please don’t do this)
- Even
Can use
=if you want
Objects and values
We will use the terms object and variable a lot when talking about code in this class
Objectsare things you save in your environment (like a set of numbers, a dataset, or a regression model)Variablerefers to columns of a data frame and to data variables that we use in models
Vectors
Think of it as a row or column in a spreadsheet
Allows same classes to be concatenated together
- Numeric
Vectors
- Character
- Factors
- Logical
Data frames
A data frame is like an Excel spreadsheet. It is two-dimensional with rows and columns.
Instead of creating a number of vectors we store all the vectors into a single DF
Can store numeric data (phone number, postal code, coordinates, etc.), float data (internet IP address, etc.), logical data (wants to receive ads: FALSE/TRUE, etc.), etc
car_model <- c("Ford Fusion", "Hyundai Accent", "Toyota Corolla")
car_price <- c(25000, 16000, 18000)
car_mileage <- c(27, 36, 32)
cars_df <- data.frame(model=car_model, price=car_price, mileage=car_mileage)
cars_df %>%
knitr::kable()| model | price | mileage |
|---|---|---|
| Ford Fusion | 25000 | 27 |
| Hyundai Accent | 16000 | 36 |
| Toyota Corolla | 18000 | 32 |
Tibbles
Modern take on data frames
Tidy data!
Each variable forms a column
Each observation forms a row
Each cell is a single measurement
Only prints few rows
Never changes your input’s type or name
Matrices
Matrices are vectors with dimensions (like a 2X5)
All the data must be the same type
Lists
While vectors are one row of data, we might want to have multiple rows or types
With a vector, it is key to understand they have to be all the same type
Lists are a grouping of variables that can be multiple types (between list items) and can be different lengths
Often function output is saved as a list for this reason
They usually have names to help you print out just a small part of the list
library(palmerpenguins)
output <- lm(flipper_length_mm ~ bill_length_mm, data = penguins)
str(output)List of 13
$ coefficients : Named num [1:2] 126.68 1.69
..- attr(*, "names")= chr [1:2] "(Intercept)" "bill_length_mm"
$ residuals : Named num [1:342] -11.766 -7.442 0.206 4.29 -3.104 ...
..- attr(*, "names")= chr [1:342] "1" "2" "3" "5" ...
$ effects : Named num [1:342] -3715.57 170.39 1.03 5.35 -2.22 ...
..- attr(*, "names")= chr [1:342] "(Intercept)" "bill_length_mm" "" "" ...
$ rank : int 2
$ fitted.values: Named num [1:342] 193 193 195 189 193 ...
..- attr(*, "names")= chr [1:342] "1" "2" "3" "5" ...
$ assign : int [1:2] 0 1
$ qr :List of 5
..$ qr : num [1:342, 1:2] -18.4932 0.0541 0.0541 0.0541 0.0541 ...
.. ..- attr(*, "dimnames")=List of 2
.. .. ..$ : chr [1:342] "1" "2" "3" "5" ...
.. .. ..$ : chr [1:2] "(Intercept)" "bill_length_mm"
.. ..- attr(*, "assign")= int [1:2] 0 1
..$ qraux: num [1:2] 1.05 1.04
..$ pivot: int [1:2] 1 2
..$ tol : num 1e-07
..$ rank : int 2
..- attr(*, "class")= chr "qr"
$ df.residual : int 340
$ na.action : 'omit' Named int [1:2] 4 272
..- attr(*, "names")= chr [1:2] "4" "272"
$ xlevels : Named list()
$ call : language lm(formula = flipper_length_mm ~ bill_length_mm, data = penguins)
$ terms :Classes 'terms', 'formula' language flipper_length_mm ~ bill_length_mm
.. ..- attr(*, "variables")= language list(flipper_length_mm, bill_length_mm)
.. ..- attr(*, "factors")= int [1:2, 1] 0 1
.. .. ..- attr(*, "dimnames")=List of 2
.. .. .. ..$ : chr [1:2] "flipper_length_mm" "bill_length_mm"
.. .. .. ..$ : chr "bill_length_mm"
.. ..- attr(*, "term.labels")= chr "bill_length_mm"
.. ..- attr(*, "order")= int 1
.. ..- attr(*, "intercept")= int 1
.. ..- attr(*, "response")= int 1
.. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. ..- attr(*, "predvars")= language list(flipper_length_mm, bill_length_mm)
.. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric"
.. .. ..- attr(*, "names")= chr [1:2] "flipper_length_mm" "bill_length_mm"
$ model :'data.frame': 342 obs. of 2 variables:
..$ flipper_length_mm: int [1:342] 181 186 195 193 190 181 195 193 190 186 ...
..$ bill_length_mm : num [1:342] 39.1 39.5 40.3 36.7 39.3 38.9 39.2 34.1 42 37.8 ...
..- attr(*, "terms")=Classes 'terms', 'formula' language flipper_length_mm ~ bill_length_mm
.. .. ..- attr(*, "variables")= language list(flipper_length_mm, bill_length_mm)
.. .. ..- attr(*, "factors")= int [1:2, 1] 0 1
.. .. .. ..- attr(*, "dimnames")=List of 2
.. .. .. .. ..$ : chr [1:2] "flipper_length_mm" "bill_length_mm"
.. .. .. .. ..$ : chr "bill_length_mm"
.. .. ..- attr(*, "term.labels")= chr "bill_length_mm"
.. .. ..- attr(*, "order")= int 1
.. .. ..- attr(*, "intercept")= int 1
.. .. ..- attr(*, "response")= int 1
.. .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
.. .. ..- attr(*, "predvars")= language list(flipper_length_mm, bill_length_mm)
.. .. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric"
.. .. .. ..- attr(*, "names")= chr [1:2] "flipper_length_mm" "bill_length_mm"
..- attr(*, "na.action")= 'omit' Named int [1:2] 4 272
.. ..- attr(*, "names")= chr [1:2] "4" "272"
- attr(*, "class")= chr "lm"Indexing
Vectors can be indexed
- 1 not 0 (👀 Python)
R as a calculator
- Typing in a simple calculation show us the result
Functions
Take arguments, do something to them, and return the result
More complex calculations can be done with functions:
- What is square root of 64?
Arguments
Some functions have settings (“arguments”) that we can adjust:
round(3.14)- Rounds off to the nearest integer (zero decimal places)
round(3.14, digits=1)- One decimal place
Getting Help
- Help files

Exercise
Open a blank new script
To paste strings together you can use the
paste() function (e.g., paste(“Hello”, “World”)). Use ?paste or Google “paste function in R” to get an idea of how to use this function.2.1 Create three vectors. One vector should have three first names. The second vector should have 3 last names. Finally, the third vector should have 3 ages. Use the
pastefunction to string together these vectors separated by a_Modify the function below and instead of returning the sum, return the mean
Outline
Why R
IDE
R commands & functions
Tidyverse & the pipe operatorReading in data
Saving R scripts
Tidyverse and pipes
- The
tidyverseis an ecosystem of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures.
Installing Tidyverse
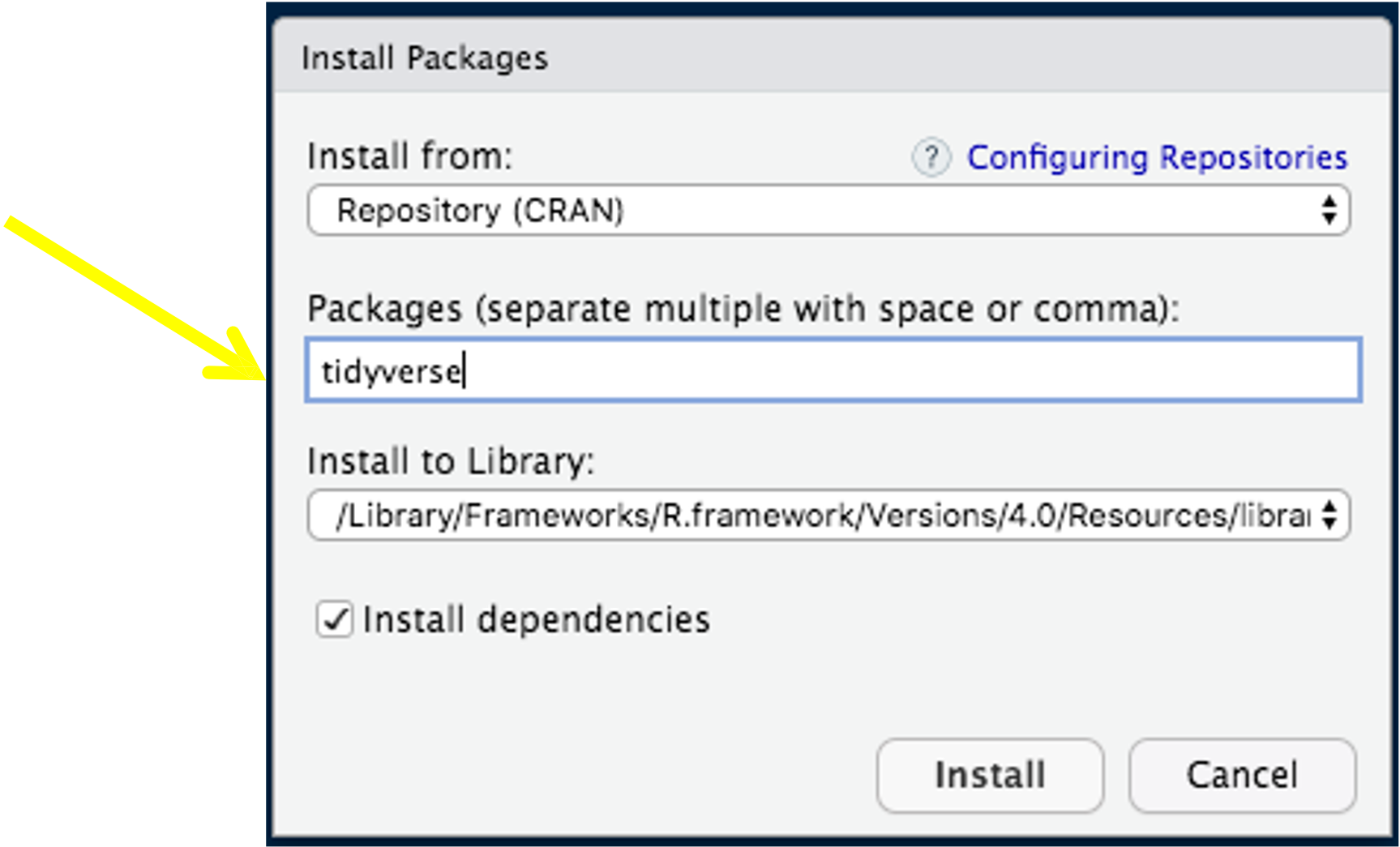
- Load package
Pipes
tidyverseprovides another interface to functionsthe pipe operator
- Makes code easier to read and follow:
This:
Can be converted into:
- Start with a and then round
|>pipe is slowly becoming more popular
Multiple functions
- Pipe operator makes it easy to do multiple functions in a row
- What is this doing?
Outline
Why R
IDE
R commands & functions
Tidyverse & the pipe operator
Reading in dataSaving R scripts
Reading in data
- Download the file

- General form:
dataframe.name <-read_csv('filename')
Reading other file types
- Excel
- SPSS
Working directories
HerepackageHerehelps set relative as opposed to absolute paths
Here
Loading the data
Note
Always create an R project before you start
Aside: naming conventions
Variable names in DFs are case-sensitive
Variable names can contain letters, numbers, underscores “_” and periods “.”
In most cases you should use snake_case to name objects
use_an_underscore_between_words
Avoid periods
Names should be short and descriptive, with descriptive being the most important feature
Janitor
Loading the data
- You can download data directly from OSF or Github
- https://osf.io/cmtxa
The faculty dataset contains aggregated data per faculty:
- faculty: Business, Economics, Political Science, Sociology
- students: number of students
- profs: number of profs
- salary: amount of salary
- costs: amount of costs dataset entails demographic and school-related information on imaginary students, such as
Load data from OSF
Looking at data
faculty students profs salary
Length:4 Min. :162.0 Min. :63.00 Min. :54246
Class :character 1st Qu.:209.2 1st Qu.:72.75 1st Qu.:56516
Mode :character Median :244.5 Median :76.50 Median :61849
Mean :247.5 Mean :73.75 Mean :65309
3rd Qu.:282.8 3rd Qu.:77.50 3rd Qu.:70642
Max. :339.0 Max. :79.00 Max. :83292
costs
Min. :24965
1st Qu.:28471
Median :31493
Mean :30370
3rd Qu.:33391
Max. :33527 Looking at data
| skim_type | skim_variable | n_missing | complete_rate | character.min | character.max | character.empty | character.n_unique | character.whitespace | numeric.mean | numeric.sd | numeric.p0 | numeric.p25 | numeric.p50 | numeric.p75 | numeric.p100 | numeric.hist |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| character | faculty | 0 | 1 | 8 | 17 | 0 | 4 | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| numeric | students | 0 | 1 | NA | NA | NA | NA | NA | 247.50 | 74.074287 | 162 | 209.25 | 244.5 | 282.75 | 339 | ▇▇▇▁▇ |
| numeric | profs | 0 | 1 | NA | NA | NA | NA | NA | 73.75 | 7.274384 | 63 | 72.75 | 76.5 | 77.50 | 79 | ▂▁▁▁▇ |
| numeric | salary | 0 | 1 | NA | NA | NA | NA | NA | 65309.00 | 13058.854084 | 54246 | 56516.25 | 61849.0 | 70641.75 | 83292 | ▇▁▃▁▃ |
| numeric | costs | 0 | 1 | NA | NA | NA | NA | NA | 30369.50 | 4023.686080 | 24965 | 28471.25 | 31493.0 | 33391.25 | 33527 | ▃▁▃▁▇ |
Looking at data
# A tibble: 4 × 5
faculty students profs salary costs
<chr> <dbl> <dbl> <dbl> <dbl>
1 Business 339 76 57273 33346
2 Economics 225 79 83292 33527
3 Political Science 264 63 66425 24965
4 Sociology 162 77 54246 29640# A tibble: 4 × 5
faculty students profs salary costs
<chr> <dbl> <dbl> <dbl> <dbl>
1 Business 339 76 57273 33346
2 Economics 225 79 83292 33527
3 Political Science 264 63 66425 24965
4 Sociology 162 77 54246 29640# A tibble: 4 × 5
faculty students profs salary costs
<chr> <dbl> <dbl> <dbl> <dbl>
1 Business 339 76 57273 33346
2 Economics 225 79 83292 33527
3 Political Science 264 63 66425 24965
4 Sociology 162 77 54246 29640Looking at data
Outline
Why RIDE
R commands, data structures, and functions
Tidyverse & the Pipe Operator
Reading in data
Manipulating dataSaving R scripts
Manipulating data
dplyris organized around verbs that manipulate data framesIsolating data:
selectfiltermutatesummarize
Select
Palmer penguins

| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year |
|---|---|---|---|---|---|---|---|
| Adelie | Torgersen | 39.1 | 18.7 | 181 | 3750 | male | 2007 |
| Adelie | Torgersen | 39.5 | 17.4 | 186 | 3800 | female | 2007 |
| Adelie | Torgersen | 40.3 | 18.0 | 195 | 3250 | female | 2007 |
| Adelie | Torgersen | NA | NA | NA | NA | NA | 2007 |
| Adelie | Torgersen | 36.7 | 19.3 | 193 | 3450 | female | 2007 |
| Adelie | Torgersen | 39.3 | 20.6 | 190 | 3650 | male | 2007 |
Select columns
The select command from dplyr allows you to subset columns matching strings:
Select helpers
Select columns
If you wanted it to be a single vector (not a tibble), use pull:
[1] male female female <NA> female male female male <NA> <NA>
[11] <NA> <NA> female male male female female male female male
[21] female male female male male female male female female male
[31] female male female male female male male female female male
[41] female male female male female male male <NA> female male
[51] female male female male female male female male female male
[61] female male female male female male female male female male
[71] female male female male female male female male female male
[81] female male female male female male male female male female
[91] female male female male female male female male female male
[101] female male female male female male female male female male
[111] female male female male female male female male female male
[121] female male female male female male female male female male
[131] female male female male female male female male female male
[141] female male female male female male male female female male
[151] female male female male female male male female female male
[161] female male female male female male female male female male
[171] female male male female female male female male <NA> male
[181] female male male female female male female male female male
[191] female male female male female male male female female male
[201] female male female male female male female male female male
[211] female male female male female male female male <NA> male
[221] female male female male male female female male female male
[231] female male female male female male female male female male
[241] female male female male female male female male male female
[251] female male female male female male <NA> male female male
[261] female male female male female male female male <NA> male
[271] female <NA> female male female male female male male female
[281] male female female male female male female male female male
[291] female male male female female male female male female male
[301] female male female male female male female male female male
[311] male female female male female male male female male female
[321] female male female male male female female male female male
[331] female male female male male female male female female male
[341] female male male female
Levels: female maleFilter
Filter
# A tibble: 6 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.3 20.6 190 3650
3 Adelie Torgersen 39.2 19.6 195 4675
4 Adelie Torgersen 38.6 21.2 191 3800
5 Adelie Torgersen 34.6 21.1 198 4400
6 Adelie Torgersen 42.5 20.7 197 4500
# ℹ 2 more variables: sex <fct>, year <int>Note, no $ or subsetting is necessary
Filter
You can have multiple logical conditions using the following:
==: equals to!: not/negation>/<: greater than / less than>=or<=: greater than or equal to / less than or equal to&: AND|: ORis.na(x): is NA!is.na(x): is not NA
Common mistakes
- Using
=instead of==
- Forgetting quotes
Exercise
- Return a df that only has NA for
sex
Mutate
Mutate
df %>%
#combines what is in species and #what is island together with #underscore
mutate(spec_island=paste(species,island, sep="_")) %>%
head()# A tibble: 6 × 9
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
# ℹ 3 more variables: sex <fct>, year <int>, spec_island <chr>Creating conditional (if-else) variables
- A general function for creating new variables based on existing variables is the
ifelse()function, which returns a value with the same shape as test which is filled with elements selected from either yes or no depending on whether the element of test isTRUEorFALSE.
Adding columns
- Mutate combined with
ifelse(condition, TRUE, FALSE), it can give you:
Case_when
Exercise
See if you can use the logical operators to manipulate our penguins data using
%>%
All of the penguin data from year 2007
All of the female penguins
Get flipper length > 150
Select only the flipper length and species columns
Summarize
Create summary statistics (mean, median, SD, sum)
- Use with
group_by
- Use with

Outline
Why R
IDE
R commands & functions
Tidyverse & the pipe operator
Reading in data
Manipulating data
Saving R scripts
Saving files
Wrapping up
You’ve learned:
Some basic programming terminology
Specific R defaults and issues
Example functions and use cases
How do I get started?
- Practice!
Helpful websites
- Google!
- Cheat sheets (https://rstudio.cloud/learn/cheat-sheets)
- Quick-R: www.statmethods.net
- R documentation: www.rdocumentation.org
- Swirl: www.swirlstats.com
- Stack Overflow: www.stackoverflow.com
- Learn Statistics with R: https://learningstatisticswithr.com/
PSY 503: Foundations of Statistics in Psychology
Comments
You can make comments on your code using the
#symbolComments are not processed by R, they provide documentation of your code for humans
Feel free to comment your personal code as much as you need to in order to understand it
Try to make your code clear enough that it can be understood even without comments